February 3rd, 2012 | 6 Comments
In the first post regarding animations we have created a bunch of png files and combined them to a single gif animation. Now we want to generate again a bunch of png files, but combine them to a movie.
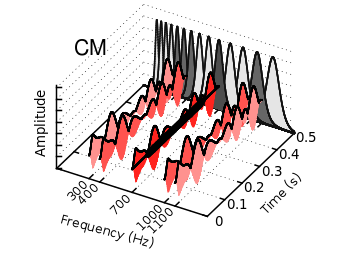
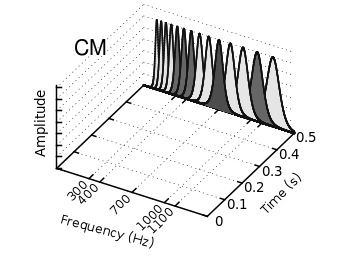
Fig. 1 Video animation of Huygens principle (code to produce this figure, loop function)
We create the png files in analogy to the gif example, hence we will discuss only the generation of the movie here. In order to compose a avi file from the png files we are using Mencoder. Gnuplot is able to directly start Mencoder by its system command.
# Create movie with mencoder ENCODER = system('which mencoder'); if (strlen(ENCODER)==0) print '=== mencoder not found ==='; exit CMD = 'mencoder mf://animation/*.png -mf fps=25:type=png -ovc lavc -lavcopts vcodec=mpeg4:mbd=2:trell -oac copy -o huygen.avi' system(CMD)
The first two lines check if Mencoder is available and quit gnuplot if not. The Mencoder command itselfs gets the directory containing the png files mf://animation/*.png, frames per second and input type-mf fps=25:type=png, video -ovc and audio -oac options, and finally of course the output file -o huygen.avi.
In order to generate a webm video file for a web site, ffmpeg can be used to convert the video.
$ ffmpeg -i huygen.avi huygen.webm