September 23rd, 2016 | 4 Comments
Suppose you have a large circular container filled with sand and measure its density at different positions. Now the goal is to display your measurements as a heat map extrapolated from your measurements, but limiting that heat map to the inner part of the container as shown in Fig. 1.
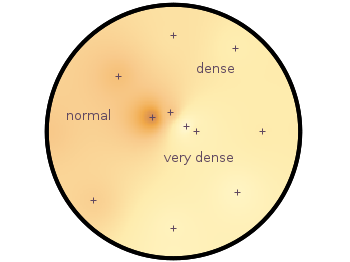
Fig. 1 Sand density measured at different positions in a circular container (code to produce this figure, sand.pal, data)
The underlying measurements are provided in the following format:
# sand_density_orig.txt #1 2 3 4 5 6 #prob x y z density description "E01" 0.00000 -1.14161 -0.020 0.7500 "dense" "E02" -0.94493 -0.81804 -0.020 0.5753 "normal" "E03" 0.75306 -0.72000 -0.020 0.7792 "dense" ...
Those data points have to be extrapolated onto a grid for the heat map, which can be achieved by the following commands.
set view map set pm3d at b map set dgrid3d 200,200,2 splot "sand_density1.txt" u 2:3:5
Fig. 2 shows the result which has two problems. The grid data is limited to the boundary given by the measurement points. In addition, the grid is always rectangular in size and not circular.
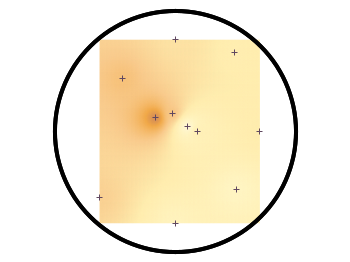
Fig. 2 Sand density measured at different positions in a circular container (code to produce this figure, sand.pal, data)
To overcome the first problem you have to add four additional points to the original data in order to stretch the grid boundary to the radius of the container. For that you have to come up with some reasonable extrapolation from the existing points. I did this in a very simple way by a mixture of linear interpolation or using the value of the nearest point. If you want to do the same with your data set you should maybe spent a little bit more effort on this.
# sand_density.txt #1 2 3 4 5 6 #prob x y z density description "E01" 0.00000 -1.14161 -0.020 0.7500 "dense" ... "xmin" -1.50000 0.00000 -0.050 0.5508 "dummy" "xmax" 1.50000 0.00000 -0.050 0.6634 "dummy" "ymin" 0.00000 -1.50000 -0.050 0.7500 "dummy" "ymax" 0.00000 1.50000 -0.050 0.6315 "dummy"
If you plot those modified data set you will get Fig. 3.
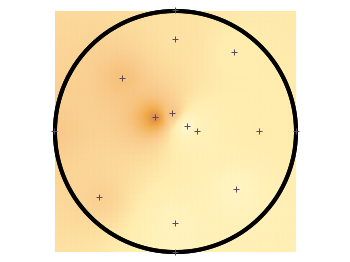
Fig. 3 Sand density measured at different positions in a circular container (code to produce this figure, sand.pal, data)
In order to limit the heat map to a circle you first extrapolate the grid using dgrid3d and store the data in a new file.
set table "tmp.txt" set dgrid3d 200,200,2 splot "sand_density2.txt" u 2:3:5 unset table
Afterwards a function is defined in order to limit the points to the inner of the circle and plot the data from the temporary file.
circle(x,y,z) = sqrt(x**2+y**2)>r ? NaN : z plot "tmp.txt" u 1:2:(circle($1,$2,$3)) w image
Finally a few labels and the original measurement points are added. The manually added points like xmin are removed by a smaller radius value. The result is then the nice circular heat map in Fig. 1.
r = 1.49 # make radius smaller to exclude interpolated edge points
set label 'normal' at -1,0.2 center front tc ls 1
set label 'dense' at 0.5,0.75 center front tc ls 1
set label 'very dense' at 0.3,-0.3 center front tc ls 1
plot "sand_density.txt" \
u (circle($2,$3,$2)):(circle($2,$3,$3)) w p ls 1